Dijkstra in R
Posted: , Last Updated:
dijkstra <- function(graph, start){
#' Implementation of dijkstra using adjacency matrix.
#' This returns an array containing the length of the shortest path from the start node to each other node.
#' It is only guaranteed to return correct results if there are no negative edges in the graph. Positive cycles are fine.
#' This has a runtime of O(|V|^2) (|V| = number of Nodes), for a faster implementation see @see ../fast/Dijkstra.java (using adjacency lists)
#' @param graph an adjacency-matrix-representation of the graph where (x,y) is the weight of the edge or 0 if there is no edge.
#' @param start the node to start from.
#' @return an array containing the shortest distances from the given start node to each other node
# This contains the distances from the start node to all other nodes
distances = rep(Inf, nrow(graph))
# This contains whether a node was already visited
visited = rep(FALSE, nrow(graph))
# The distance from the start node to itself is of course 0
distances[start] = 0
# While there are nodes left to visit...
repeat{
# ... find the node with the currently shortest distance from the start node...
shortest_distance = Inf
shortest_index = -1
for(i in seq_along(distances)) {
# ... by going through all nodes that haven't been visited yet
if(distances[i] < shortest_distance && !visited[i]){
shortest_distance = distances[i]
shortest_index = i
}
}
cat("Visiting node ", shortest_index, " with current distance ", shortest_distance, "\n")
if(shortest_index == -1){
# There was no node not yet visited --> We are done
return (distances)
}
# ...then, for all neighboring nodes that haven't been visited yet....
for(i in seq_along(graph[shortest_index,])) {
# ...if the path over this edge is shorter...
if(graph[shortest_index,i] != 0 && distances[i] > distances[shortest_index] + graph[shortest_index,i]){
# ...Save this path as new shortest path.
distances[i] = distances[shortest_index] + graph[shortest_index,i]
cat("Updating distance of node ", i, " to ", distances[i], "\n")
}
# Lastly, note that we are finished with this node.
visited[shortest_index] = TRUE
cat("Visited nodes: ", visited, "\n")
cat("Currently lowest distances: ", distances, "\n")
}
}
}
About the algorithm and language used in this code snippet:
Dijkstra's Algorithm
The Dijkstra algorithm is an algorithm used to solve the shortest path problem in a graph. This means that given a number of nodes and the edges between them as well as the “length” of the edges (referred to as “weight”), the Dijkstra algorithm is finds the shortest path from the specified start node to all other nodes. Nodes are sometimes referred to as vertices (plural of vertex) - here, we’ll call them nodes.
Description of the Algorithm
The basic principle behind the Dijkstra algorithm is to iteratively look at the node with the currently smallest distance to the source and update all not yet visited neighbors if the path to it via the current node is shorter. In more detail, this leads to the following Steps:
- Initialize the distance to the starting node as 0 and the distances to all other nodes as infinite
- Set all nodes to “unvisited”
-
While we haven’t visited all nodes:
- Find the node with currently shortest distance from the source (for the first pass, this will be the source node itself)
- For all nodes next to it that we haven’t visited yet, check if the currently smallest distance to that neighbor is bigger than if we were to go via the current node
- If it is, update the smallest distance of that neighbor to be the distance from the source to the current node plus the distance from the current node to that neighbor
In the end, the array we used to keep track of the currently shortest distance from the source to all other nodes will contain the (final) shortest distances.
Example of the Algorithm
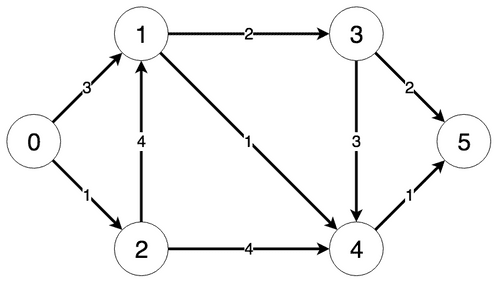
The steps the algorithm performs on this graph if given node 0 as a starting point, in order, are:
- Visiting node 0
- Updating distance of node 1 to 3
- Updating distance of node 2 to 1
- Visited nodes: 0
- Currently lowest distances: [0, 3, 1, Infinite, Infinite, Infinite]
-
Visiting node 1 with current distance 1
- Updating distance of node 3 to 5
- Visited nodes: 0, 2
- Currently lowest distances: [0, 3, 1, 5, Infinite, Infinite]
-
Visiting node 3 with current distance 3
- Updating distance of node 4 to 4
- Visited nodes: 0, 1, 2
- Currently lowest distances: [0, 3, 1, 5, 4, Infinite]
-
Visiting node 4 with current distance 4
- Updating distance of node 5 to 5
- Visited nodes: 0, 1, 2, 4
- Currently lowest distances: [0, 3, 1, 5, 4, 5]
-
Visiting node 5 with current distance 5
- No distances to update
- Visited nodes: 0, 1, 2, 3, 4
- Currently lowest distances: [0, 3, 1, 5, 4, 5]
-
Visiting node 5 with current distance 5
- No distances to update
- Visited nodes: 0, 1, 2, 3, 4, 5
All nodes visited Final lowest distances: [0, 3, 1, 5, 4, 5]
Runtime of the Algorithm
The runtime complexity of Dijkstra depends on how it is implemented. If a min-heap is used to determine the next node to visit, and adjacency is implemented using adjacency lists, the runtime is O(|E| + |V|log|V|) (|V| = number of Nodes, |E| = number of Edges). If a we simply search all distances to find the node with the lowest distance in each step, and use a matrix to look up whether two nodes are adjacent, the runtime complexity increases to O(|V|^2).
Space of the Algorithm
The space complexity of Dijkstra depends on how it is implemented as well and is equal to the runtime complexity.
R
R is an interpreted language first released in 1993 with a significant increase in popularity in recent years. It is primarily used for data mining and -science as well as statistics, and is a popular language in non-computer science disciplines ranging from Biology to Physics. R is dynamically typed, and has one of the widest variety of libraries for statistics, machine learning, data mining etc.
Getting to “Hello World” in R
The most important things first - here’s how you can run your first line of code in R.
- Download and install the latest version of R from r-project.org. You can also download an earlier version if your use case requires it.
- Open a terminal, make sure the
Rcommand is working, and that the command your’re going to be using is referring to the version you just installed by runningR --version. If you’re getting a “command not found” error (or similar), try restarting your command line, and, if that doesn’t help, your computer. If the issue persists, here are some helpful StackOverflow questions for Windows, Mac and Linux. - As soon as that’s working, you can run the following snippet:
print("Hello World"). You have two options to run this: 3.1 RunRin the command line, just paste the code snippet and press enter (PressCTRL + Dand typenfollowed by enter to exit). 3.2 Save the snippet to a file, name it something ending with.R, e.g.hello_world.R, and runRscript hello_world.R. Tip: use thelscommand (dirin Windows) to figure out which files are in the folder your command line is currently in.
That’s it! Notice how printing something to the console is just a single line in R - this low entry barrier and lack of required boilerplate code is a big part of the appeal of R.
Fundamentals in R
To understand algorithms and technologies implemented in R, one first needs to understand what basic programming concepts look like in this particular language.
Variables and Arithmetic
Variables in R are really simple, no need to declare a datatype or even declare that you’re defining a variable; R knows this implicitly. R is also able to easily define objects and their property, in multiple different ways.
some_value = 10
my_object <- list(my_value = 4)
attr(my_object, 'other_value') <- 3
print((some_value + my_object$my_value + attr(my_object, 'other_value'))) # Prints 17Arrays
Working with arrays is similarly simple in R:
# Create 2 vectors of length 3
vector1 <- c(1,2,3)
vector2 <- c(4,5,6)
# Create names for rows and columns (optional)
column.names <- c("column_1","column_2","column_3")
row.names <- c("row_1","row_2")
# Concatenate the vectors (as rows) to form an array, providing dimensions and row/column names
result <- array(c(vector1,vector2), dim = c(2,3), dimnames = list(row.names, column.names))
print(result)
# Prints:
# column_1 column_2 column_3
# row_1 1 3 5
# row_2 2 4 6As those of you familiar with other programming language like Java might have already noticed, those are not native arrays, but rather lists dressed like arrays. This means that arrays in R are considerably slower than in lower level programming languages. This is a trade off R makes in favor of simplicity. There are, however, packages which implement real arrays that are considerably faster.
Conditions
Just like most programming languages, R can do if-else statements:
value = 1
if(value==1){
print("Value is 1")
} else if(value==2){
print("Value is 2")
} else {
print("Value is something else")
}R can also do switch statements, although they are implemented as a function, unlike in other languages like Java:
x <- switch(
1,
"Value is 1",
"Value is 2",
"Value is 3"
)
print(x)Note that this function is pretty useless, but there are other functions for more complex use cases.
Loops
R supports both for and while loops as well as break and next statements (comparable to continue in other languages).
Additionally, R supports repeat-loops, which are comparable to while(true) loops in other languages, but simplify the code a little bit.
value <- 0
repeat {
value <- value + 1
if(value > 10) {
break
}
}
print(value)
value <- 0
while (value <= 10) {
value = value + 1
}
print(value)
value <- c("Hello","World","!")
for ( i in value) {
print(i)
}
for(i in 1:10){
print(i)
}Functions
Functions in R are easily defined and, for better or worse, do not require specifying return or arguments types. Optionally, a default for arguments can be specified:
my_func <- function (
a = "World"
) {
print(a)
return("!")
}
my_func("Hello")
print(my_func())(This will print “Hello”, “World”, and then ”!“)
Syntax
R requires the use of curly brackets ({}) to surround code blocks in conditions, loops, functions etc.;
While this can lead to some annoying syntax errors, it also means the use of whitespace for preferred formatting (e.g. indentation of code pieces) does not affect the code.
Advanced Knowledge of R
For more information, R has a great Wikipedia article. The official website is r-project.org.